File:Tiger phylogenetic relationships.png
From Vigyanwiki
Tiger phylogenetic relationships.png
Size of this preview: 574 × 600 pixels. Other resolutions: 230 × 240 pixels | 460 × 480 pixels | 735 × 768 pixels | 980 × 1,024 pixels | 2,001 × 2,090 pixels.
Original file (2,001 × 2,090 pixels, file size: 684 KB, MIME type: image/png)
This file is from Wikimedia Commons and may be used by other projects. The description on its file description page there is shown below.
Summary
| DescriptionTiger phylogenetic relationships.png |
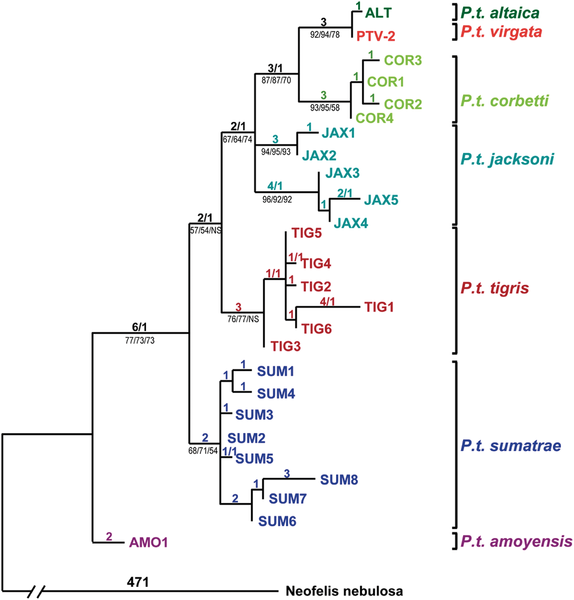
English: Haplotype designations are color coded by subspecies of the tigers that carried them. PTV-2 is a Caspian tiger (Panthera tigris virgata) specimen for which all gene segments attempted (1257 bp) in Caspian tigers were successfully sequenced. Other Caspian tigers produced partial sequences identical to PTV-2. The only exceptions were three individuals, each displaying a single derived nucleotide difference when compared to PTV-2 (found only in that individual and in no other tigers of any subspecies). Likewise, the only mtDNA haplotype carried by Amur or “Siberian” tigers (P. t. altaica) proved to be a single derived step away from the haplotype of PTV-2, suggesting a close relationship between the Amur and Caspian tiger subspecies. Tiger haplotypes carried by all but the Caspian subspecies are from a previously published dataset, while a clouded leopard (Neofelis nebulosa) sequence (GenBank DQ257669) was used to root the tree. The tree depicted was inferred using maximum parsimony, with the number of steps/homoplasies listed above the branches, while (for major clades) bootstrap percentages are listed below branches for maximum parsimony, maximum likelihood and Neighbour Joining methods. We used full length mtDNA sequences of clouded leopard, leopard and snow leopard to root the tree; all combinations of 1, 2 or 3 outgroups yielded trees with similar topology to the one depicted, with the same basal position for the P. t. amoyensis AMO1 haplotype, and a close relationship between P.t. virgata and P. t. altaica haplotypes. doi:info:doi/10.1371/journal.pone.0004125.g002 |
||
| Date | |||
| Source | Driscoll CA, Yamaguchi N, Bar-Gal GK, Roca AL, Luo S, et al. (2009) Mitochondrial Phylogeography Illuminates the Origin of the Extinct Caspian Tiger and Its Relationship to the Amur Tiger. PLoS ONE 4(1): e4125. doi:10.1371/journal.pone.0004125 | ||
| Author | Driscoll CA, Yamaguchi N, Bar-Gal GK, Roca AL, Luo S, et al. | ||
| Permission (Reusing this file) |
|
Captions
Add a one-line explanation of what this file represents
Items portrayed in this file
depicts
14 January 2009
700,586 byte
2,090 pixel
2,001 pixel
image/png
19f07b91d4e2594a77d4f1b6c393fb9bd37184f0
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 16:11, 26 February 2012 |  | 2,001 × 2,090 (684 KB) | wikimediacommons>Pmaas |
File usage
The following page uses this file:
Metadata
This file contains additional information, probably added from the digital camera or scanner used to create or digitise it.
If the file has been modified from its original state, some details may not fully reflect the modified file.
| Author |
|
|---|---|
| Horizontal resolution | 118.11 dpc |
| Vertical resolution | 118.11 dpc |
| Software used |
|
